Research
Multicellular organisms also need to control their use of cellular energy stores. Glucose metabolism plays a crucial role in organismal homeostasis, influencing energy consumption, cell proliferation, stress resistance and lifespan. Defective glucose utilization causes numerous diseases ranging from diabetes to an increased tendency to develop tumors. For cells to respond appropriately to changes in energy status, they need a fine-tune system to modulate chromatin dynamics in order to respond to metabolic cues. Reciprocally, chromatin changes necessary for cellular functions need as well to be coupled to metabolic adaptations.
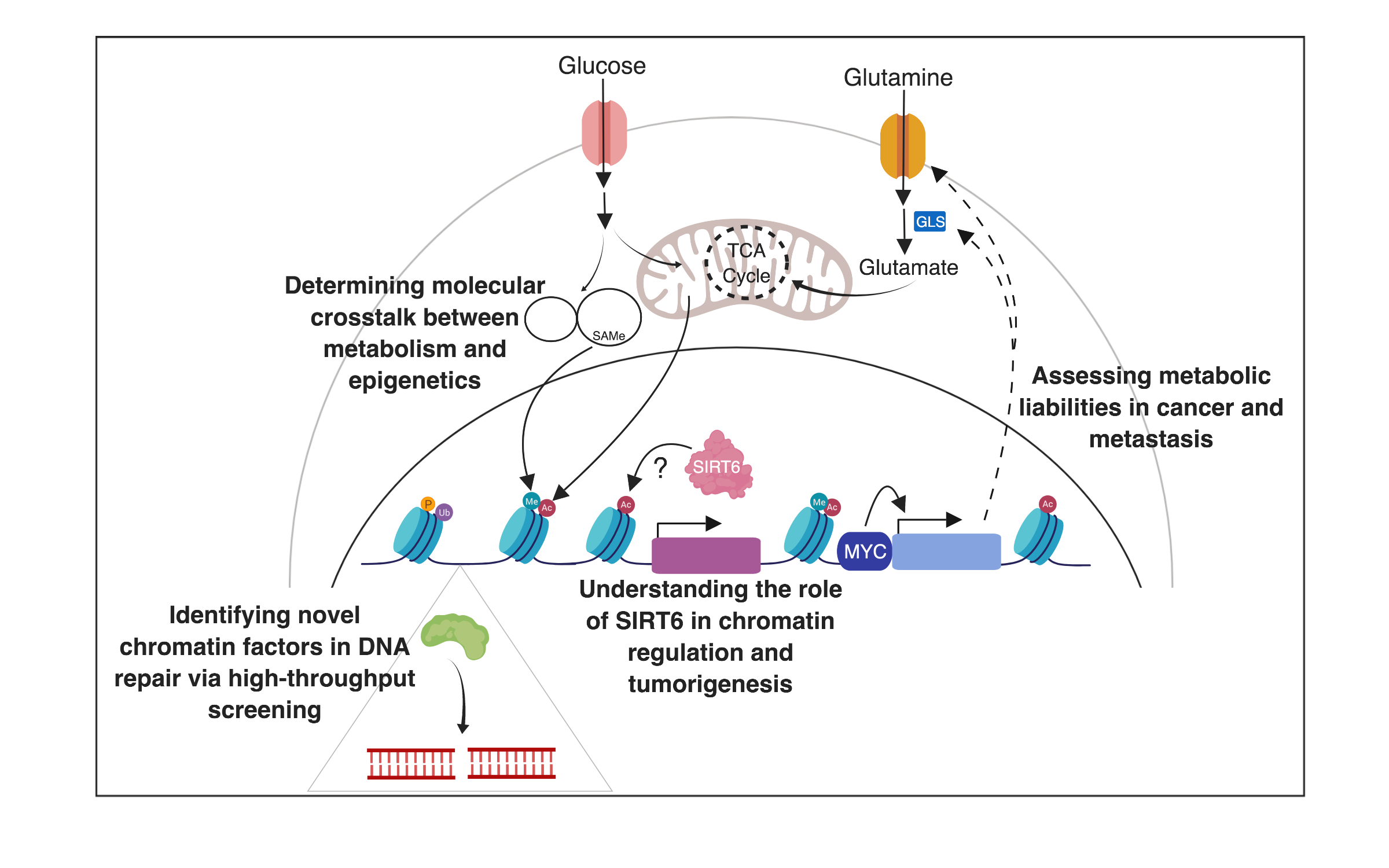
Our lab is interested in understanding the influence of chromatin on nuclear processes (gene transcription, DNA recombination and DNA repair) and the relationship between chromatin dynamics and the metabolic adaptation of cells. One of our interests is on the study of a group of proteins called SIRTs, the mammalian homologues of the yeast Sir2. Sir2 is a chromatin silencer that functions as an NAD-dependent histone deacetylase to inhibit DNA transcription and recombination. In the past few years, we have been exploring the crosstalk between epigenetics and metabolism. In particular, our work has focused on the mammalian Sir2 homologue, SIRT6. In recent years, we have identified SIRT6 as a key modulator of metabolism. Mice lacking SIRT6 exhibit severe metabolic defects, including hypoglycemia and hypoinsulinemia. SIRT6 appears to modulate glucose flux inside the cells, functioning as a histone H3K9 deacetylase to silence glycolytic genes acting as a co-expressor of Hif1alpha, in this way directing glucose away from to reduce intracellular ROS levels. This function appears critical for glucose homeostasis, as SIRT6 deficient animals die early in life from hypoglycemia. Remarkably, SIRT6 acts as a tumor suppressor in colon cancer, regulating cancer metabolism through mechanisms that by-pass known oncogenic pathways. Cancer cells prefer fermentation (i.e., lactate production) to respiration. Despite being described by biochemist and Nobel laureate Otto Warburg decades ago (i.e., the Warburg effect), the molecular mechanisms behind this metabolic switch remain a mystery. We believe SIRT6 may function as a critical modulator of the Warburg effect, providing a long-sought molecular explanation to this phenomenon. We have also uncovered key roles for SIRT6 in DNA repair (anchoring the chromatin remodeler SNF2H to DNA breaks) and early development (acting as a repressor of pluripotent genes), indicating broad biological functions for this chromatin deacetylase. More recently, we identified SIRT6 as a robust tumor suppressor in pancreatic cancer, where it silences the oncofetal protein Lin28b, protecting against aggressive tumor phenotypes. As such, SIRT6 represents an example of a chromatin factor modulated by cancer cells to acquire “epigenetic plasticity”. Our current studies are directed at determining how the DNA repair and metabolic functions of SIRT6 may be related to each other.
In addition, we more recently started expanding our research. We are exploring novel metabolic liabilities in cancer, including assessing the ability of cells to adapt to extreme nutrient conditions. On the other hand, we are looking into broader chromatin roles in DNA repair, using unique combinations of chromatin libraries and DNA repair assays where we can perform high throughput screenings.
Specific Projects:
- Deciphering how SIRT6 regulates chromatin structure
- Determining the role of SIRT6 in tumorigenesis using mouse models
- Elucidating the role of histone modifications and chromatin dynamics in DNA repair
- Determining molecular crosstalk between epigenetics and metabolism
- Assessing metabolic liabilities in cancer and metastases